step | name | version |
|---|---|---|
Adapter trimming | dorado | v0.6.1 |
Primer trimming | cutadapt | v4.6 |
ITS region extraction | ITSxpress | v2.0.1 |
Quality filtering | chopper | v0.7.0 |
Chimera detection | VSEARCH | v2.21.1 |
Subsampling reads | seqtk | v1.4 |
Dimension reduction | UMAP | v0.4.6 |
Clustering | HDBSCAN | v0.8.26 |
Taxonomic assignment | dnabarcoder | v1.0.6 |
Fastq/fasta file manipulation | seqkit | v2.6.1 |
Draft read selection | fastANI | v1.34 |
Read mapping | minimap | v2.28 |
Read polishing | racon | v1.5.0 |
Read polishing | medaka | v1.12.0 |
11 Appendices
1 Workflow
2 Mock scenarios
Barcode | Sample | Number of raw reads | Number of reads after QC |
|---|---|---|---|
25 | Puccinia striiformis var tritici | 38,761 | 9304 (24.0%) |
26 | Puccinia triticina (Puccinia recondita)e,u | 21,731 | 2216 (10.2%) |
27 | Puccinia graminis | 24,352 | 6994 (28.7%) |
28 | Austropuccinia psidii | 28,916 | 8378 (29.0%) |
29 | Leptosphaeria maculans | 40,207 | 7123 (17.7%) |
30 | Sclerotinia sclerotiorum | 30,922 | 5461 (17.7%) |
31 | Botrytis cinerea | 35,580 | 5953 (16.7%) |
32 | Botrytis fabae | 28,674 | 4533 (15.8%) |
33 | Zymoseptoria tritici | 31,584 | 2747 (8.7%) |
34 | Sporisorium scitamineum | 29,141 | 3610 (12.4%) |
35 | Erysiphe necator | 50,836 | 4889 (9.6%) |
36 | Austropuccinia psidiiu | 29,523 | 8299 (28.1%) |
37 | Pyrenophora tritici-repentis | 51,246 | 5655 (11.0%) |
38 | Cryptovalsa ampelina | 37,861 | 4816 (12.7%) |
39 | Eutypa lata | 25,259 | 2886 (11.4%) |
40 | Diplodia seriata | 39,280 | 4821 (12.3%) |
41 | Fusarium pseudograminearum | 37,072 | 5716 (15.4%) |
42 | Aspergillus flavum (Aspergillus flavus) | 52,132 | 2775 (5.3%) |
43 | Aspergillus fumigatus | 48,170 | 4829 (10.0%) |
44 | Aspergillus nigere,u | 106,560 | 352 (0.3%) |
45 | Blastobotrys adeninivorans | 56,595 | 11158 (19.7%) |
46 | Blastobotrys proliferans | 45,388 | 10511 (23.2%) |
47 | Candida albicans | 35,572 | 6889 (19.4%) |
48 | candida boletica (Candida boleticola) | 45,525 | 10374 (22.8%) |
49 | Candida caryicola | 42,127 | 9482 (22.5%) |
50 | Candida catenulata (Diutina catenulata) | 70,924 | 15285 (21.6%) |
51 | Candida dublinsiensis (Candida dubliniensis) | 62,732 | 12240 (19.5%) |
52 | Candida glabrata (Nakaseomyces glabratus) | 48,252 | 5206 (10.8%) |
53 | Candida metapsilosis | 48,123 | 11117 (23.1%) |
54 | Candida orthopsilosis | 45,771 | 10222 (22.3%) |
55 | Candida parapsilosis | 53,561 | 10754 (20.1%) |
56 | Candida tropicalis | 48,486 | 10111 (20.9%) |
57 | Candid zeylanoides (Candida zeylanoides) | 47,125 | 11643 (24.7%) |
58 | Cryptococcus albidus (Naganishia albida)e,u | 262 | 6 (2.3%) |
59 | Cryptococcus gattil VG I (Cryptococcus gattii VG I) | 47,998 | 9161 (19.1%) |
60 | Cryptococcus gattii VG IIIu | 21,127 | 3817 (18.1%) |
61 | Cryptococcus neoformans VNIu | 19,970 | 2842 (14.2%) |
62 | Cryptococcus neoformans VN IIII | 54,596 | 10852 (19.9%) |
63 | Fusarium proliferatume,u | 79,693 | 4894 (6.1%) |
64 | Galactomyces geotrichum (Geotrichum candidum)e,u | 15,962 | 12 (0.1%) |
65 | Geotrichum candidum | 82,477 | 26203 (31.8%) |
66 | Kluyveromyces lactis | 70,007 | 14285 (20.4%) |
67 | Kluyveromyces marxianus | 70,959 | 9212 (13.0%) |
68 | Kodamaea ohmeri | 65,595 | 11929 (18.2%) |
69 | Meyerozyma guillermondii (Meyerozyma guilliermondii)e,u | 29,370 | 12 (0.0%) |
70 | Nakazawaea ernobil (Nakazawaea ernobii) | 69,285 | 15320 (22.1%) |
71 | Penicillium chrysogenum | 64,025 | 3569 (5.6%) |
72 | Pichia kudriavzevii | 56,683 | 9807 (17.3%) |
73 | Pichia membranifacien (Pichia membranifaciens) | 39,163 | 2963 (7.6%) |
74 | Rhodotorula mucilaginosa | 53,680 | 8580 (16.0%) |
75 | Scedosporium auranticum (Scedosporium aurantiacum) | 52,475 | 4348 (8.3%) |
76 | Scedosporium boydii | 43,418 | 4766 (11.0%) |
77 | Trichomonascus ciferrii | 39,999 | 5338 (13.3%) |
78 | Trichosporon asahil (Trichosporon asahii) | 49,679 | 11219 (22.6%) |
79 | Wickerhamomyces anomalus | 45,298 | 8840 (19.5%) |
80 | Yamadazyma mexicana | 44,496 | 10733 (24.1%) |
81 | Yamadazyma scolyti | 44,166 | 8874 (20.1%) |
82 | Yarrowia lipolyticae,u | 45,485 | 10 (0.0%) |
83 | Diaporthe sp CCL067 | 36,374 | 5055 (13.9%) |
84 | Quambalaria cyanescens CCL055 | 49,988 | 6172 (12.3%) |
85 | Entoleuca sp CCL052 | 41,661 | 3845 (9.2%) |
86 | Cortinarius globuliformis CM4 | 57,951 | 7732 (13.3%) |
87 | Asteroma sp CCL068 | 45,777 | 6254 (13.7%) |
88 | Asteroma sp CCL060 | 42,061 | 4449 (10.6%) |
89 | Tuber brumale | 102,909 | 15169 (14.7%) |
e,Excluded from even abundance scenario | |||
uExcluded from uneven abundance scenario | |||
step | name | version | command |
|---|---|---|---|
Basecalling + | guppy | v6.4.2 | guppy_basecaller \ |
2.1 Clustering - NanoCLUST
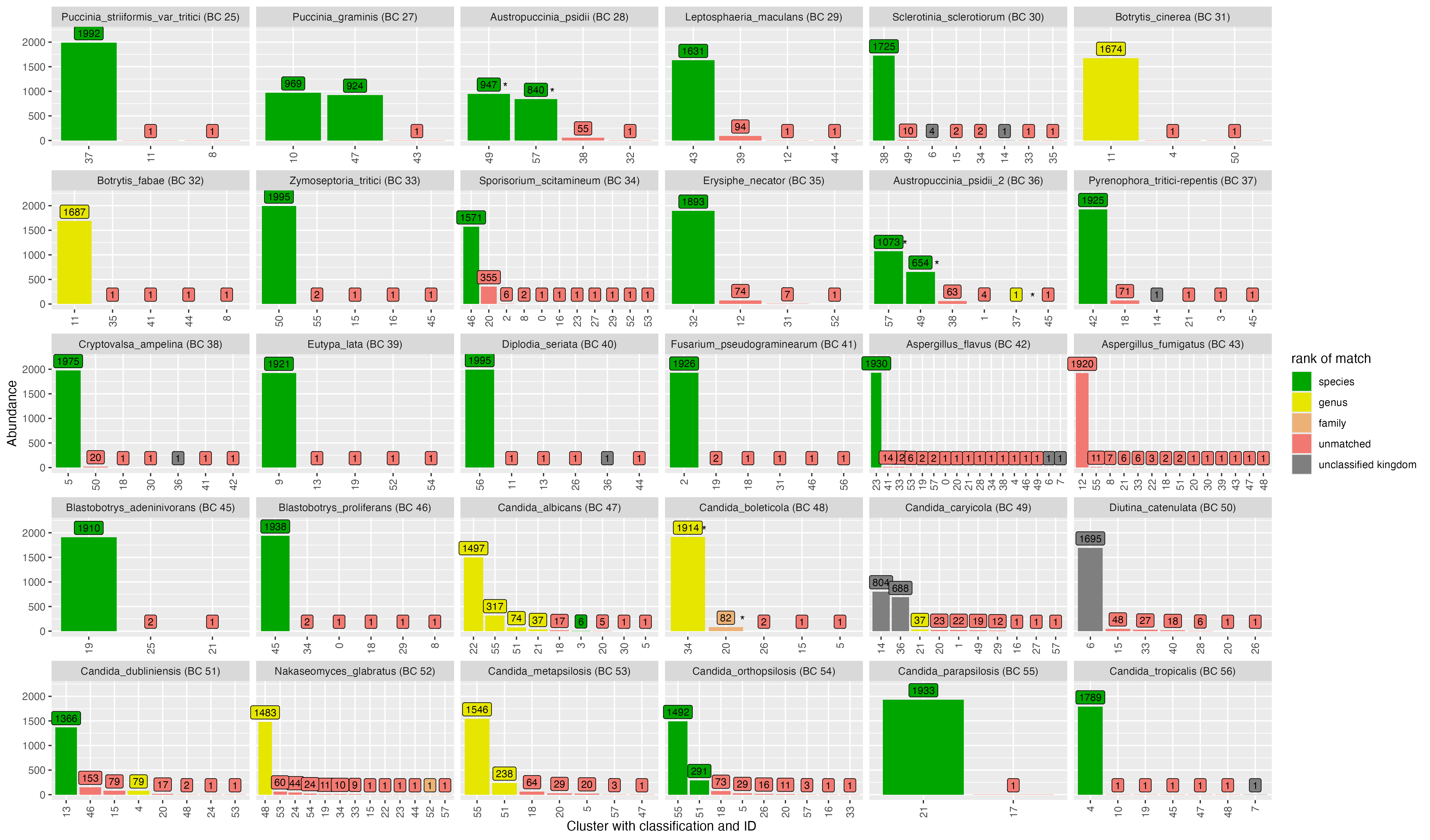
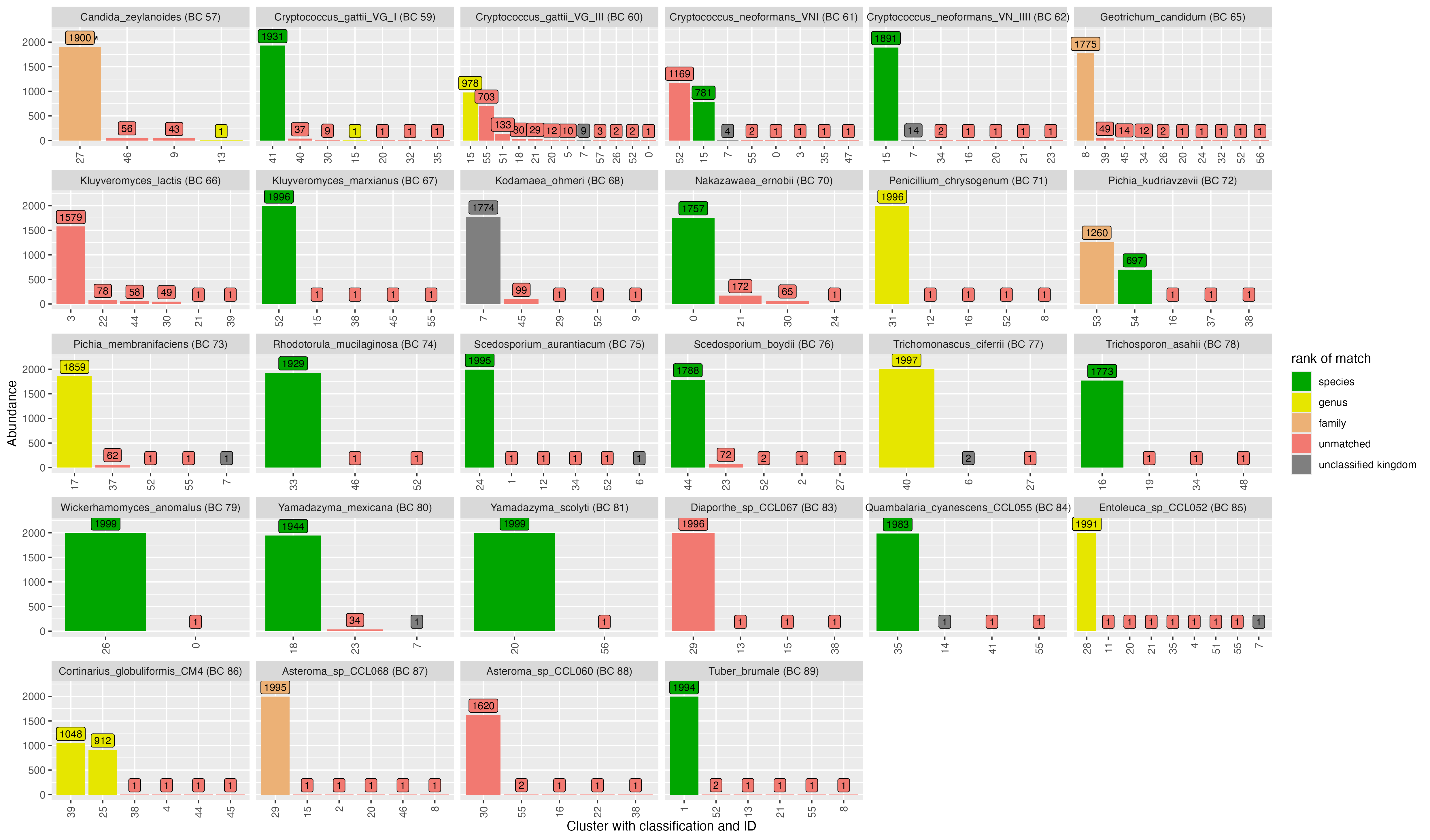
OTU ID | genus | species | score | confidence |
|---|---|---|---|---|
0 | Nakazawaea | Nakazawaea ernobii | 100.00% | 83.87% |
1 | Tuber | Tuber brumale | 99.88% | 90.68% |
2 | Fusarium | Fusarium pseudograminearum | 100.00% | 82.32% |
3 | Candida | Candida albicans | 100.00% | 92.48% |
4 | Candida | Candida tropicalis | 100.00% | 92.48% |
5 | Cryptovalsa | Cryptovalsa ampelina | 100.00% | 82.32% |
6 | unclassified kingdom | unclassified kingdom | 78.00% | 0.00% |
7 | unclassified kingdom | unclassified kingdom | 79.25% | 0.00% |
8 | Dipodascus | Dipodascus capitatus | 90.50% | 90.87% |
9 | Eutypa | Eutypa lata | 100.00% | 93.14% |
10 | Puccinia | Puccinia graminis | 100.00% | 82.32% |
11 | Botrytis | Botrytis genus | 100.00% | 74.10% |
12 | Inopinatum | Inopinatum lactosum | 100.00% | 82.32% |
13 | Candida | Candida dubliniensis | 100.00% | 92.48% |
14 | unclassified kingdom | unclassified kingdom | 86.00% | 0.00% |
15 | Cryptococcus | Cryptococcus neoformans | 100.00% | 93.32% |
16 | Trichosporon | Trichosporon asahii | 100.00% | 82.32% |
17 | Pichia | Pichia genus | 95.75% | 82.49% |
18 | Yamadazyma | Yamadazyma mexicana | 100.00% | 86.96% |
19 | Blastobotrys | Blastobotrys adeninivorans | 100.00% | 95.61% |
20 | Yamadazyma | Yamadazyma scolyti | 100.00% | 86.96% |
21 | Candida | Candida parapsilosis | 100.00% | 92.48% |
22 | Candida | Candida africana | 100.00% | 92.48% |
23 | Aspergillus | Aspergillus flavus | 100.00% | 90.31% |
24 | Scedosporium | Scedosporium aurantiacum | 100.00% | 82.32% |
25 | Cortinarius | Cortinarius genus | 99.50% | 57.27% |
26 | Wickerhamomyces | Wickerhamomyces anomalus | 100.00% | 96.38% |
27 | Debaryomycetaceae family | Debaryomycetaceae family | 100.00% | 73.25% |
28 | Entoleuca | Entoleuca genus | 100.00% | 57.27% |
29 | Gnomoniopsis | Gnomoniopsis paraclavulata | 100.00% | 95.16% |
30 | Diaporthe | Diaporthe foeniculina | 100.00% | 82.32% |
31 | Penicillium | Penicillium rubens | 100.00% | 90.71% |
32 | Erysiphe | Erysiphe necator | 100.00% | 89.16% |
33 | Rhodotorula | Rhodotorula mucilaginosa | 100.00% | 82.32% |
34 | Kurtzmaniella | Kurtzmaniella genus | 100.00% | 73.25% |
35 | Quambalaria | Quambalaria cyanescens | 100.00% | 82.32% |
36 | unclassified kingdom | unclassified kingdom | 87.50% | 0.00% |
37 | Puccinia | Puccinia striiformis | 100.00% | 82.32% |
38 | Sclerotinia | Sclerotinia sclerotiorum | 100.00% | 96.69% |
39 | Cortinarius | Cortinarius genus | 99.67% | 57.27% |
40 | Trichomonascus | Trichomonascus genus | 100.00% | 57.27% |
41 | Cryptococcus | Cryptococcus gattii | 100.00% | 93.32% |
42 | Pyrenophora | Pyrenophora tritici-repentis | 100.00% | 93.15% |
43 | Leptosphaeria | Leptosphaeria maculans | 100.00% | 91.09% |
44 | Scedosporium | Scedosporium boydii | 100.00% | 82.32% |
45 | Blastobotrys | Blastobotrys proliferans | 100.00% | 95.61% |
46 | Sporisorium | Sporisorium scitamineum | 100.00% | 90.16% |
47 | Puccinia | Puccinia graminis | 100.00% | 82.32% |
48 | Nakaseomyces | Nakaseomyces genus | 100.00% | 80.41% |
49 | Puccinia | Puccinia psidii | 100.00% | 82.32% |
50 | Zymoseptoria | Zymoseptoria tritici | 100.00% | 82.32% |
51 | Candida | Candida orthopsilosis | 100.00% | 92.48% |
52 | Kluyveromyces | Kluyveromyces marxianus | 100.00% | 82.32% |
53 | Komagataella | Komagataella pastoris | 100.00% | 82.32% |
54 | Pichia | Pichia kudriavzevii | 100.00% | 86.66% |
55 | Candida | Candida orthopsilosis | 99.76% | 92.48% |
56 | Diplodia | Diplodia seriata | 100.00% | 93.98% |
57 | Puccinia | Puccinia psidii | 100.00% | 82.32% |
2.2 Clustering - VSEARCH
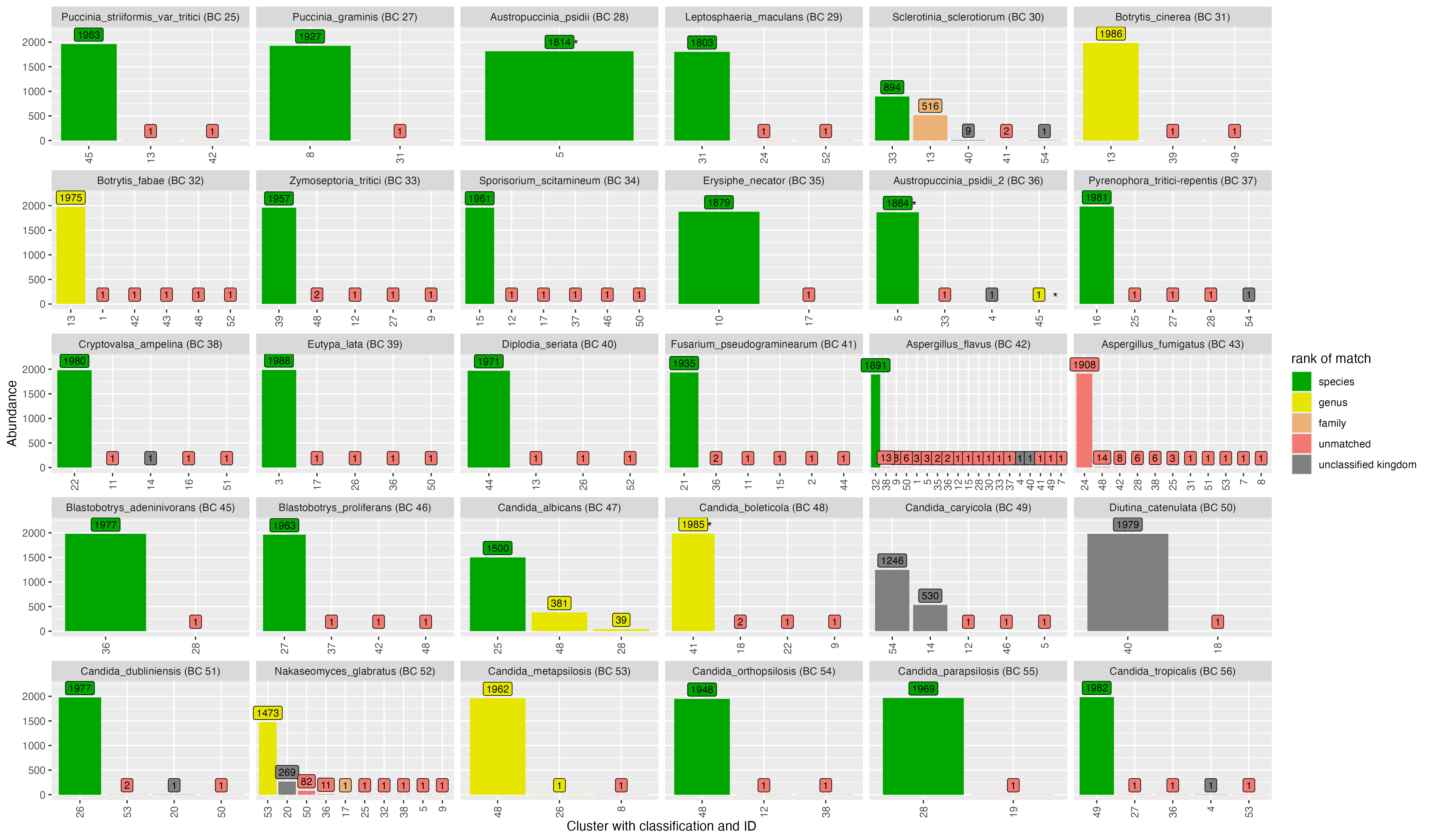
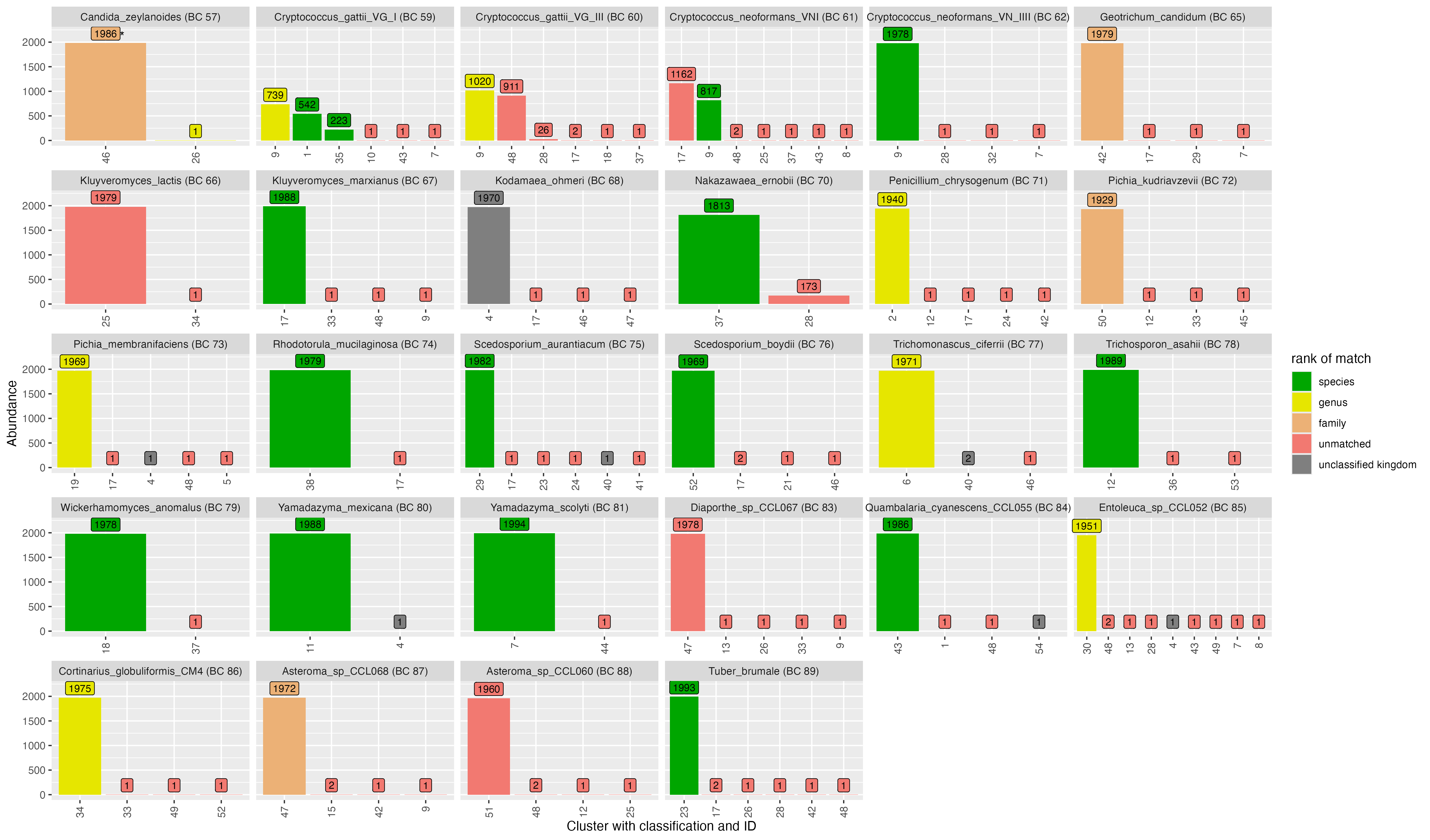
OTU ID | genus | species | score | confidence |
|---|---|---|---|---|
1 | Cryptococcus | Cryptococcus gattii | 99.88% | 90.68% |
2 | Penicillium | Penicillium rubens | 100.00% | 82.32% |
3 | Eutypa | Eutypa lata | 100.00% | 92.48% |
4 | unclassified kingdom | unclassified kingdom | 100.00% | 92.48% |
5 | Puccinia | Puccinia psidii | 100.00% | 82.32% |
6 | Trichomonascus | Trichomonascus genus | 78.00% | 0.00% |
7 | Yamadazyma | Yamadazyma scolyti | 79.25% | 0.00% |
8 | Puccinia | Puccinia graminis | 90.50% | 90.87% |
9 | Cryptococcus | Cryptococcus neoformans | 100.00% | 93.14% |
10 | Erysiphe | Erysiphe necator | 100.00% | 82.32% |
11 | Yamadazyma | Yamadazyma mexicana | 100.00% | 74.10% |
12 | Trichosporon | Trichosporon asahii | 100.00% | 82.32% |
13 | Botrytis | Botrytis genus | 100.00% | 92.48% |
14 | unclassified kingdom | unclassified kingdom | 86.00% | 0.00% |
15 | Sporisorium | Sporisorium scitamineum | 100.00% | 93.32% |
16 | Pyrenophora | Pyrenophora tritici-repentis | 100.00% | 82.32% |
17 | Kluyveromyces | Kluyveromyces marxianus | 95.75% | 82.49% |
18 | Wickerhamomyces | Wickerhamomyces anomalus | 100.00% | 86.96% |
19 | Pichia | Pichia genus | 100.00% | 95.61% |
20 | unclassified kingdom | unclassified kingdom | 100.00% | 86.96% |
21 | Fusarium | Fusarium pseudograminearum | 100.00% | 92.48% |
22 | Cryptovalsa | Cryptovalsa ampelina | 100.00% | 92.48% |
23 | Tuber | Tuber brumale | 100.00% | 90.31% |
24 | Inopinatum | Inopinatum lactosum | 100.00% | 82.32% |
25 | Candida | Candida albicans | 99.50% | 57.27% |
26 | Candida | Candida dubliniensis | 100.00% | 96.38% |
27 | Blastobotrys | Blastobotrys proliferans | 100.00% | 73.25% |
28 | Candida | Candida parapsilosis | 100.00% | 57.27% |
29 | Scedosporium | Scedosporium aurantiacum | 100.00% | 95.16% |
30 | Entoleuca | Entoleuca genus | 100.00% | 82.32% |
31 | Leptosphaeria | Leptosphaeria maculans | 100.00% | 90.71% |
32 | Aspergillus | Aspergillus flavus | 100.00% | 89.16% |
33 | Sclerotinia | Sclerotinia sclerotiorum | 100.00% | 82.32% |
34 | Cortinarius | Cortinarius genus | 100.00% | 73.25% |
35 | Cryptococcus | Cryptococcus gattii | 100.00% | 82.32% |
36 | Blastobotrys | Blastobotrys adeninivorans | 87.50% | 0.00% |
37 | Nakazawaea | Nakazawaea ernobii | 100.00% | 82.32% |
38 | Rhodotorula | Rhodotorula mucilaginosa | 100.00% | 96.69% |
39 | Zymoseptoria | Zymoseptoria tritici | 99.67% | 57.27% |
40 | unclassified kingdom | unclassified kingdom | 100.00% | 57.27% |
41 | Kurtzmaniella | Kurtzmaniella genus | 100.00% | 93.32% |
42 | Dipodascus | Dipodascus capitatus | 100.00% | 93.15% |
43 | Quambalaria | Quambalaria cyanescens | 100.00% | 91.09% |
44 | Diplodia | Diplodia seriata | 100.00% | 82.32% |
45 | Puccinia | Puccinia striiformis | 100.00% | 95.61% |
46 | Debaryomycetaceae family | Debaryomycetaceae family | 100.00% | 90.16% |
47 | Gnomoniopsis | Gnomoniopsis paraclavulata | 100.00% | 82.32% |
48 | Candida | Candida orthopsilosis | 100.00% | 80.41% |
49 | Candida | Candida tropicalis | 100.00% | 82.32% |
50 | Komagataella | Komagataella pastoris | 100.00% | 82.32% |
51 | Diaporthe | Diaporthe foeniculina | 100.00% | 92.48% |
52 | Scedosporium | Scedosporium boydii | 100.00% | 82.32% |
53 | Nakaseomyces | Nakaseomyces genus | 100.00% | 82.32% |
54 | unclassified kingdom | unclassified kingdom | 100.00% | 86.66% |
3 Real world - Soil case study
Sample | Number of raw reads | Number of reads after QC |
|---|---|---|
AL1 | 239,709 | 186363 (77.7%) |
AL2 | 231,117 | 152282 (65.9%) |
AL3 | 186,404 | 137353 (73.7%) |
AL4 | 118,352 | 91943 (77.7%) |
AL5 | 73,496 | 53703 (73.1%) |
AL6 | 147,981 | 93942 (63.5%) |
AP1 | 71,005 | 51227 (72.1%) |
AP2 | 103,684 | 69442 (67.0%) |
AP3 | 97,940 | 69235 (70.7%) |
AP4 | 114,140 | 84609 (74.1%) |
AP5 | 144,789 | 90968 (62.8%) |
AP6 | 3,577 | 1173 (32.8%) |
EC1 | 29,232 | 17466 (59.7%) |
EC2 | 72,957 | 55584 (76.2%) |
EC3 | 122,122 | 85317 (69.9%) |
EC5 | 111,698 | 72093 (64.5%) |
EC6 | 104,171 | 79082 (75.9%) |
EV1 | 98,575 | 73681 (74.7%) |
EV2 | 76,320 | 51884 (68.0%) |
EV3 | 66,069 | 49847 (75.4%) |
EV4 | 94,882 | 64587 (68.1%) |
EV5 | 111,569 | 70090 (62.8%) |
EV6 | 215,888 | 138717 (64.3%) |
ExCon | 334,396 | 6672 (2.0%) |
PCRNegCon | 71,377 | 2400 (3.4%) |
step | name | version | command |
|---|---|---|---|
Basecalling | dorado | v0.7.1 | |
Demultiplexing | minibar | v0.25 | minibar.py -F \ |
classification | count | score | cutoff | ReferenceID | closest match |
|---|---|---|---|---|---|
unclassified kingdom | 239,241 | 90.06% | KC152167 | Geastrum genus | |
Archaeorhizomyces genus | 41,602 | 95.86% | 95.60% | UDB01263360 | Archaeorhizomyces genus |
Tomentella genus | 38,361 | 99.83% | 95.60% | UDB06025650 | Tomentella genus |
unclassified kingdom | 37,311 | 93.75% | GU222314 | Amanita australis | |
Laccaria genus | 35,455 | 99.83% | 95.60% | OQ064943 | Laccaria genus |
unclassified kingdom | 34,909 | 87.87% | KP889944 | Archaeorhizomycetes class | |
Mortierella genus | 33,161 | 100.00% | 95.60% | UDB04902775 | Mortierella genus |
Clavulina genus | 32,208 | 100.00% | 95.60% | DQ672317 | Clavulina genus |
Scleroderma genus | 25,070 | 98.74% | 95.60% | UDB06676397 | Scleroderma genus |
unclassified kingdom | 23,785 | 88.16% | UDB04632058 | Piloderma genus | |
unclassified kingdom | 22,597 | 92.49% | DQ309136 | Rozellomycota phylum | |
Hydnodontaceae family | 21,074 | 94.53% | 93.80% | OL828784 | Trechispora praefocata |
Oidiodendron genus | 17,240 | 100.00% | 95.60% | OK584555 | Oidiodendron genus |
Leucoagaricus genus | 16,640 | 99.51% | 95.60% | UDB03049341 | Leucoagaricus genus |
unclassified kingdom | 16,578 | 49.28% | UDB0760917 | Inocybe genus | |
unclassified kingdom | 16,310 | 48.77% | UDB0343814 | Helotiales order | |
unclassified kingdom | 15,946 | 90.83% | DQ474677 | Piloderma genus | |
Penicillium genus | 15,849 | 100.00% | 95.30% | UDB04263718 | Penicillium genus |
Amanita genus | 15,823 | 96.85% | 95.60% | GU222314 | Amanita australis |
unclassified kingdom | 15,541 | 93.17% | UDB01262630 | Archaeorhizomyces genus | |
Aspergillus felis | 11,912 | 100.00% | 99.20% | OQ152598 | Aspergillus felis |
Tomentella genus | 10,887 | 100.00% | 95.60% | UDB06165394 | Tomentella genus |
unclassified kingdom | 9,990 | 58.30% | UDB0756057 | Sebacina genus | |
unclassified kingdom | 9,824 | 36.89% | UDB0757809 | Pyronemataceae family | |
Talaromyces genus | 9,195 | 100.00% | 93.70% | OP497912 | Talaromyces genus |
unclassified kingdom | 8,996 | 58.30% | UDB0756057 | Sebacina genus | |
unclassified kingdom | 8,761 | 0.00% | |||
Penicillium genus | 8,737 | 100.00% | 95.30% | UDB03940467 | Penicillium genus |
Inocybe subferruginea | 8,729 | 99.37% | 99.10% | KP636874 | Inocybe subferruginea |
unclassified kingdom | 8,547 | 93.27% | UDB0761652 | Cortinarius genus |
classification | count | score | cutoff | ReferenceID | closest match |
|---|---|---|---|---|---|
Penicillium genus | 14,879 | 100.00% | 95.30% | UDB03808858 | Penicillium genus |
unclassified kingdom | 3,191 | 90.24% | KC152167 | Geastrum genus | |
Laccaria genus | 2,143 | 99.83% | 95.60% | UDB02413463 | Laccaria genus |
unclassified kingdom | 2,069 | 93.75% | GU222314 | Amanita australis | |
Mortierella genus | 1,904 | 100.00% | 95.60% | UDB04902775 | Mortierella genus |
unclassified kingdom | 1,816 | 90.42% | UDB02700124 | Geastrum genus | |
Archaeorhizomyces genus | 1,418 | 95.86% | 95.60% | UDB01263360 | Archaeorhizomyces genus |
unclassified kingdom | 1,263 | 90.06% | KC152167 | Geastrum genus | |
unclassified kingdom | 1,219 | 92.49% | DQ309136 | Rozellomycota_gen_Incertae_sedis genus | |
Oidiodendron genus | 1,036 | 100.00% | 95.60% | OK584555 | Oidiodendron genus |
Penicillium citrinum | 1,023 | 100.00% | 99.20% | MN783029 | Penicillium citrinum |
Penicillium genus | 960 | 100.00% | 95.30% | UDB04263718 | Penicillium genus |
Clavulina genus | 905 | 100.00% | 95.60% | DQ672317 | Clavulina genus |
Scleroderma genus | 846 | 98.74% | 95.60% | UDB06676397 | Scleroderma genus |
unclassified kingdom | 842 | 49.28% | UDB0760917 | Inocybe genus | |
Aspergillus felis | 827 | 100.00% | 99.20% | OQ152598 | Aspergillus felis |
Tomentella genus | 810 | 100.00% | 95.60% | UDB06171495 | Tomentella genus |
Leucoagaricus genus | 636 | 99.51% | 95.60% | UDB03049341 | Leucoagaricus genus |
Amanita genus | 629 | 96.85% | 95.60% | GU222314 | Amanita australis |
Talaromyces genus | 614 | 100.00% | 93.70% | OP497912 | Talaromyces genus |
Hydnodontaceae family | 588 | 94.53% | 93.80% | OL828784 | Trechispora praefocata |
Leucoagaricus genus | 579 | 99.67% | 95.60% | UDB03049341 | Leucoagaricus genus |
Penicillium genus | 571 | 100.00% | 95.30% | OQ870808 | Penicillium genus |
unclassified kingdom | 566 | 90.45% | UDB06347141 | Trechispora genus | |
Hydnodontaceae family | 556 | 94.73% | 93.80% | OL828784 | Trechispora praefocata |
Hydnaceae family | 548 | 95.04% | 93.80% | UDB07049378 | Sistotrema genus |
unclassified kingdom | 512 | 93.27% | UDB0761652 | Cortinarius genus | |
unclassified kingdom | 500 | 90.83% | DQ474677 | Piloderma genus | |
unclassified kingdom | 465 | 90.66% | KC152167 | Geastrum genus | |
Meyerozyma guilliermondii | 440 | 100.00% | 99.10% | OR761551 | Meyerozyma guilliermondii |